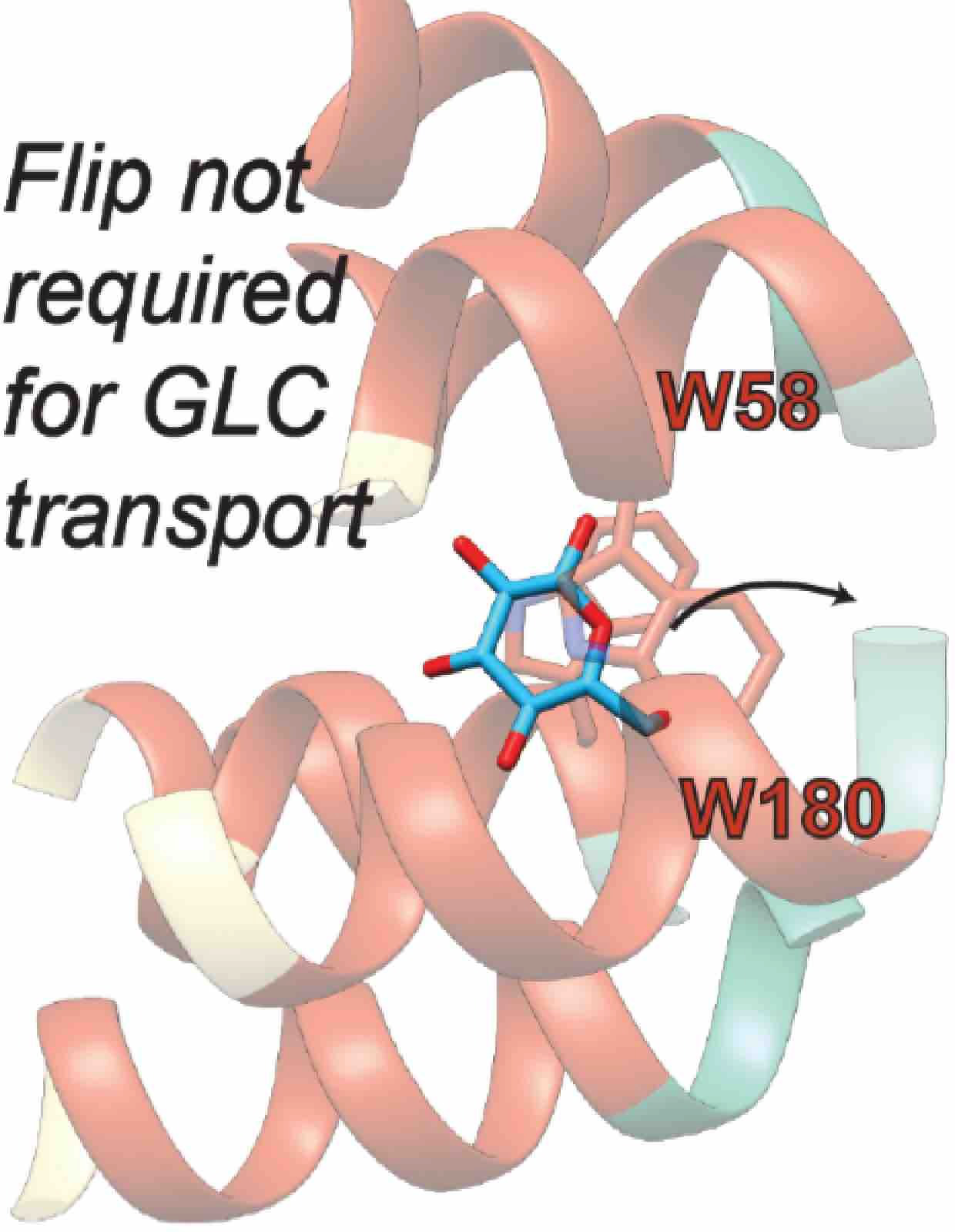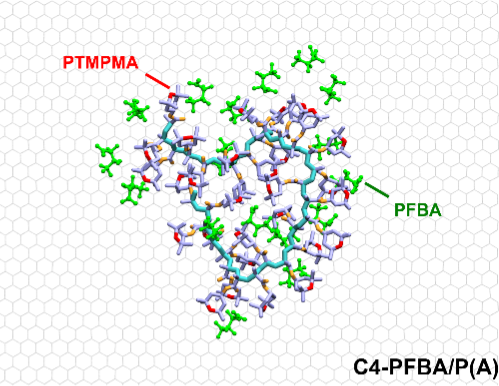
Imparting selective fluorophilic interactions in redox copolymers for the electrochemically mediated capture of short-chain perfluoroalkyl substances
Journal of American Chemical Society, In press, 2023.
Anaira Román Santiago, Song Yin, Johannes Elbert, Jiho Lee, Diwakar Shukla, and Xiao Su*
BibTeX Anaira Román Santiago, Song Yin, Johannes Elbert, Jiho Lee, Diwakar Shukla, and Xiao Su*
BibTeX

Thirty years of molecular dynamics simulations on post-translational modifications of proteins
Physical Chemistry Chemical Physics, Volume 24, Pages 26371-26397, 2022.
Austin T Weigle, Jiangyan Feng and Diwakar Shukla*
Publisher's website
BibTeX Austin T Weigle, Jiangyan Feng and Diwakar Shukla*
BibTeX

Integration of machine learning with computational structural biology of plants
Biochemical Journal, Volume 479, Issue 8, Pages 921-928, 2022.
Jiming Chen and Diwakar Shukla*
BibTeX Jiming Chen and Diwakar Shukla*
BibTeX
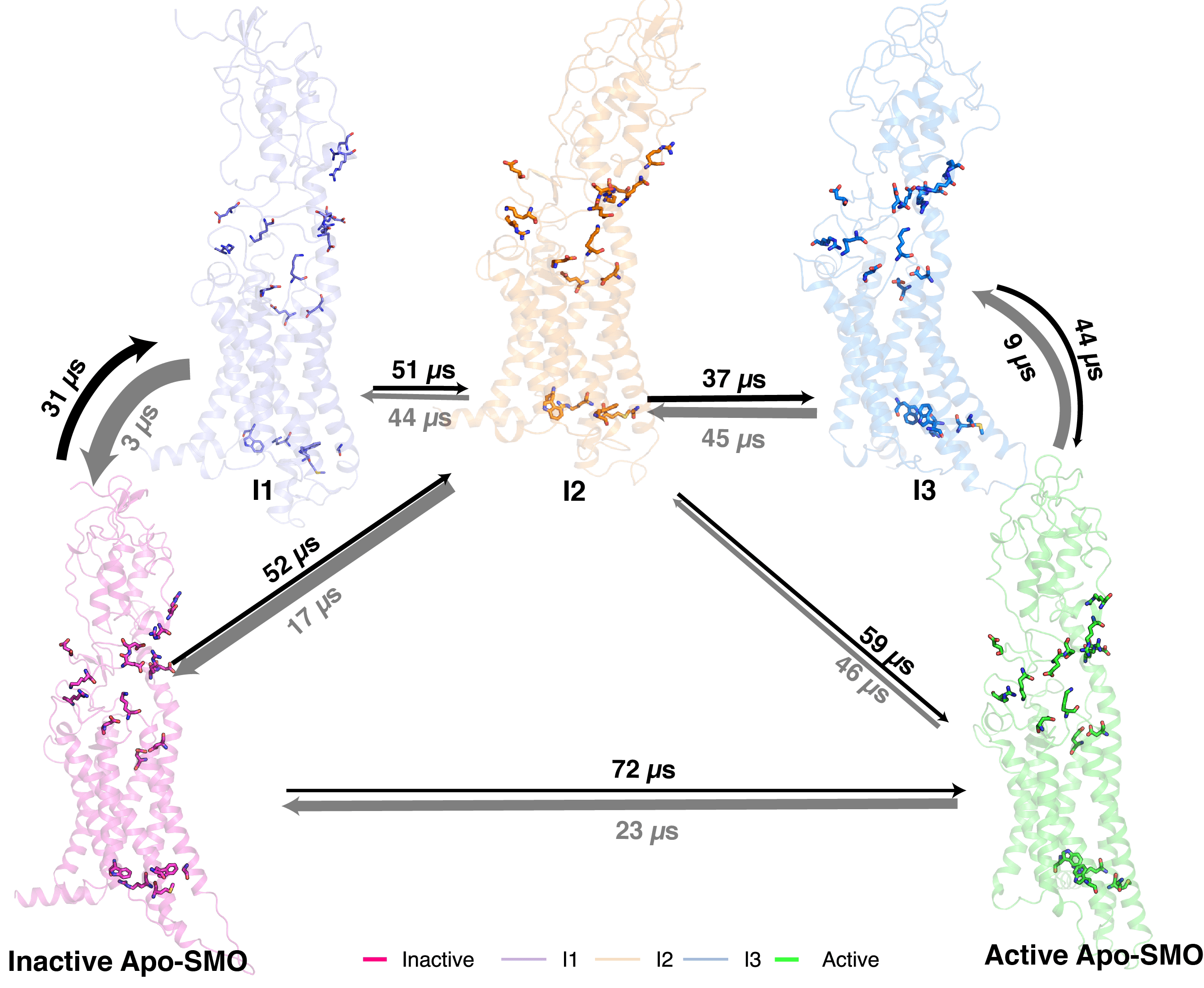
Activation Mechanism of the Human Smoothened Receptor
Biophysical Journal, In press, 2023.
Prateek D. Bansal, Soumajit Dutta and Diwakar Shukla
Publisher's website
BibTeX Prateek D. Bansal, Soumajit Dutta and Diwakar Shukla
BibTeX
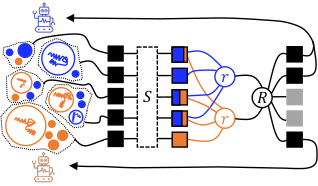
Multiagent Reinforcement Learning-Based Adaptive Sampling for Conformational Dynamics of Proteins
Journal of Chemical Theory and Computation, Volume 18, Issue 9, Pages 5422-5434, 2022.
Diego E. Kleiman and Diwakar Shukla*
Publisher's website
BibTeX Diego E. Kleiman and Diwakar Shukla*
BibTeX

Active Learning of the Conformational Ensemble of Proteins using Maximum Entropy VAMPNets
bioRxiv, doi:10.1101/2023.01.12.523801
Diego E Kleiman and Diwakar Shukla
Publisher's website
BibTeX Diego E Kleiman and Diwakar Shukla
BibTeX
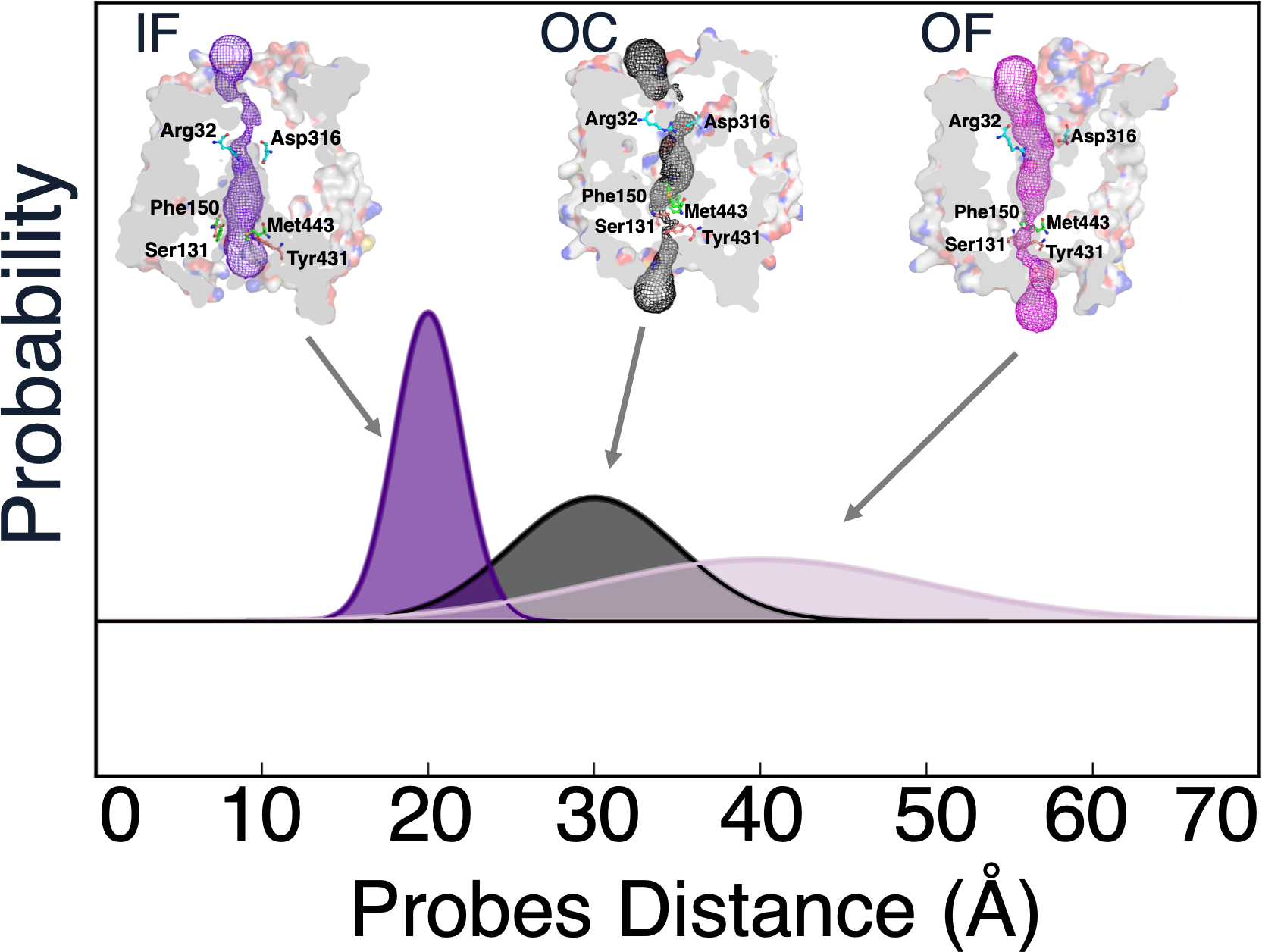
Reconciling Membrane Protein Simulations with Experimental DEER Spectroscopy Data
Physical Chemistry Chemical Physics, In press, 2023.
Mittal, Shriyaa and Shukla, Diwakar
Publisher's website
BibTeX Mittal, Shriyaa and Shukla, Diwakar
BibTeX
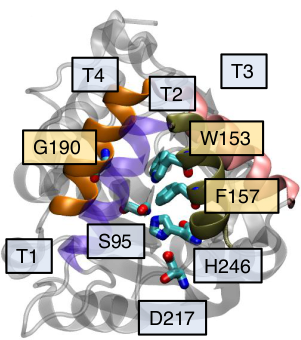
Mechanistic Basis for Enhanced Strigolactone Sensitivity in KAI2 Triple Mutant
bioRxiv, doi: 10.1101/2023.01.18.524622
Briana L Sobecks, Jiming Chen and Diwakar Shukla
Publisher's website
BibTeX Briana L Sobecks, Jiming Chen and Diwakar Shukla
BibTeX
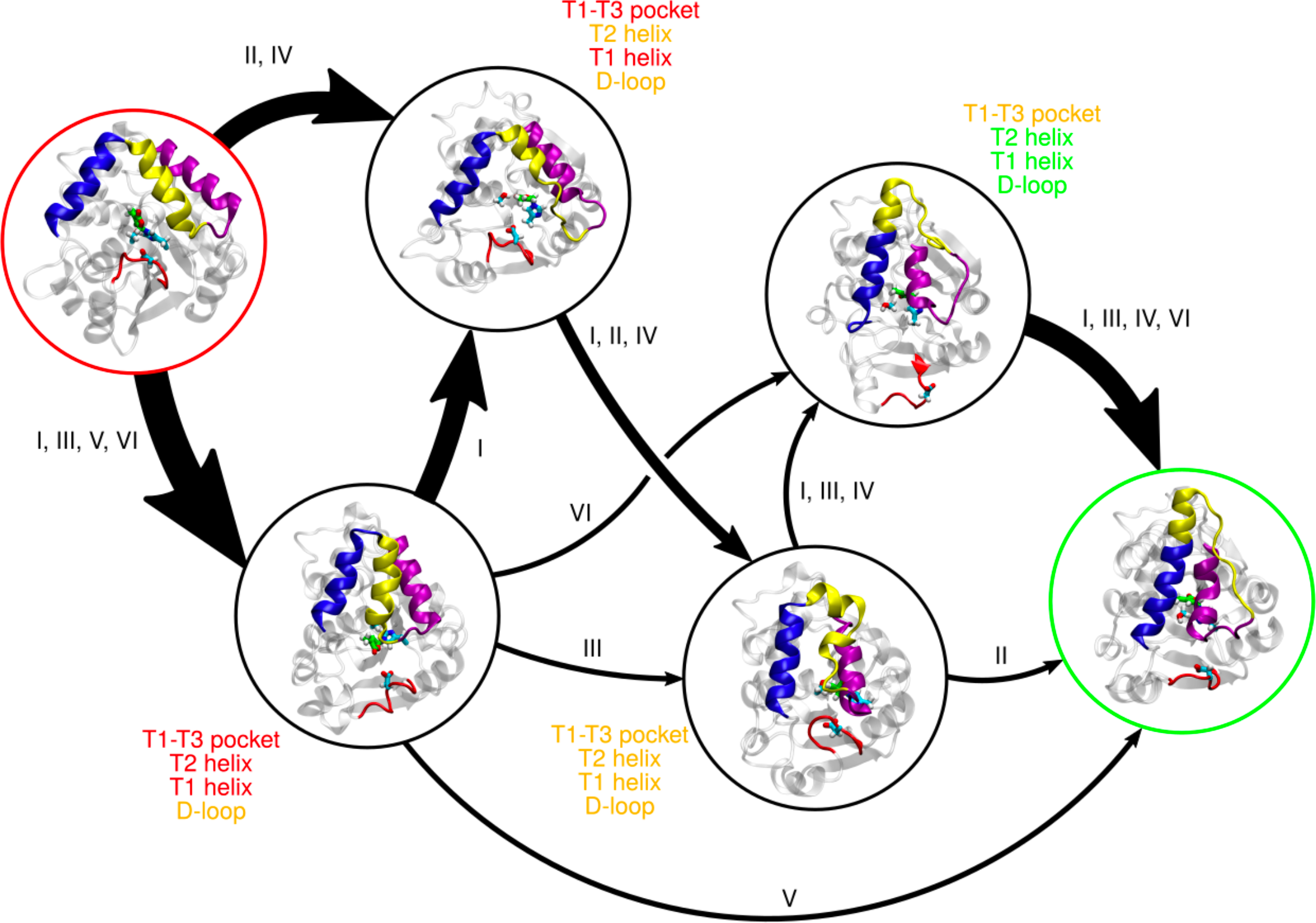
Effect of Histidine Covalent Modification on Strigolactone Receptor Activation and Selectivity.
Biophysical Journal, In press, 2023.
Jiming Chen and Diwakar Shukla
Publisher's website
BibTeX Jiming Chen and Diwakar Shukla